
Dr. Girisha Katta
Department: Department of Medical Genetics
Kasturba Medical College, Manipal
Manipal Academy of Higher Education
Manipal-576104
Contact: +91 820 2923149
E-mail: girish.katta@manipal.edu
Orcid ID: https://orcid.org/0000-0002-0139-8239
Website: https://manipal.edu/kmc-manipal/department-faculty/faculty-list/girisha-km.html
Education:
· MBBS, Government Medical College, Mysore, 1992-1998
· Diploma in Child Health, College of Physicians and Surgeons of Bombay, 2001
· MD (Pediatrics), Seth GS Medical College and KEM Hospitals, Mumbai, 1999-2002
· DM (Medical Genetics), Sanjay Gandhi Postgraduate Institute of Medical Sciences, Lucknow, 2003-2005
· Fellow, Cedar Sinai Medical Centre, Los Angeles
Research Achievements:
Since the last 15 years, Dr Girisha has been using genomic approaches to understand human genetic disorders. He has contributed to discovery of new genetic disorders and determining clinical, genomic and mutation profiles of several rare genetic disorders in India. He played a vital role in research, patient care, education and development of this recent medical specialty in our country.
Dr Girisha’s clinical practice and research have resulted in improved diagnosis and understanding of genetic alterations associated with rare diseases. These include progressive pseudorheumatoid dysplasia, Morquio syndrome A, GM1 gangliosidosis, multicentric osteolysis, nodulosis and arthropathy, pyknodysostosis, Mucopolysaccharidoses types I and II, Niemann-Pick disease, Tay-Sachs disease, Larsen syndrome and Waardenburg syndrome. Dr Girisha is one of the first physician-scientists to utilize new genomic technologies to unravel hitherto undescribed rare diseases and their genetic basis in India. These efforts began with identification of a human ciliopathy affecting the skeleton, eyes and brain caused by mutations in IFT52 gene. His team soon described a new syndrome of skeletal dysplasia with joint dislocations and discovered that itwas caused by mutations in EXOC6B. This was followed by identification of a new syndrome intwo children with intellectual disability, ataxia and facial dysmorphism due to mutations in EBF3.
Dr Girisha and his team are credited with discovery of two new childhood neurodevelopmental disorders in quick succession. The first one affected mitochondrial iron-sulphur cluster biogenesis pathway due to a mutation in ISCA1 gene that led to a very severe neuroregressive disorder in five children. He played a crucial role in discovery of a childhood progressive neurodevelopmental disorder with microcephaly, seizures, and spastic quadriparesis that resulted from defects in AIMP2 gene. Similar strategies were applied to identify a new skeletal dysplasia with large epiphyses caused by alterations in PISD gene and followed it by elucidating the mechanism of the disease. Since the year 2000, up to 50 genes for rare diseases have been identified with major contributions from Indian researchers. Dr. Girisha and his colleagues described nearly twenty of them in the last five years emphasizing his contributions to the field of disease-gene discovery.
Online Mendelian Inheritance in Man (OMIM) currently catalogues eleven of them as new monogenic disorders. These include Spondyloepimetaphyseal dysplasia with joint laxity, type 3 (#618395), Leukodystrophy, hypomyelinating, 17 (#618006), Multiple mitochondrial dysfunctions syndrome 5 (#617613), Laurin-Sandrow syndrome (#135750), Hypotonia, ataxia, and delayed development syndrome (#617330), Arthrogryposis, distal with impaired proprioception and touch (#617146), Short-rib thoracic dysplasia 16 with or without polydactyly (#617102), Shukla-Vernon syndrome (#301029), Spondyloepimetaphyseal dysplasia, X-linked (#300106), Spondyloepimetaphyseal dysplasia, SPONASTRIME type (#271510) and Anauxetic dysplasia 3 (#618853).
Dr Girisha has a special interest in skeletal dysplasia as evidenced by ten of his newly described disease-gene associations appended to this group. As an expert in this area globally, he earned a place in the nosology group of International Skeletal Dysplasia Society that is tasked with recognition and classification of genetic disorders of the skeleton. As evident from his CV, Dr Girisha has published over 200 peer-reviewed articles including many in high impact (IF) publications.
In addition to research and clinical practice, Dr Girisha is a pioneer in Medical Genetics education in India. He established the first Master of Science program in Genetic Counseling in India at Manipal that is now emulated in other centers. He is a supervisor for 18 PhD scholars (8 as a co-supervisor) including two clinician-scientists. He also started one of four Doctor of Medicine (DM) courses in Medical Genetics in India. He is one of the founding members of Society for Indian Academy of Medical Genetics (www.iamg.in ) and spearheads its publication Genetic Clinics. He also plays a huge role in advocating genetics and genetic testing in the region through local television, newspapers and magazines and talks to laypersons.
Dr Girisha founded the department of Medical Genetics at our institute and in just 5 years he built a formidable team of young scientists at Manipal to arguably become one of the largest center for clinical genetics in India. He work also had global impact on rare disease research, diagnosis and care of patients with rare diseases and medical genetics education.
Dr. Rama Rao Damerla
Department of Medical Genetics
Kasturba Medical College Manipal
Manipal Academy of Higher Education

Research Experience
Dr. Rama Rao Damerla joined Manipal Academy of Higher Education in June 2019, as an Assistant Professor and Government of India, Department of Biotechnology – Ramalingaswami Fellow in the Department of Medical Genetics. He graduated with a PhD in Human Genetics in 2011 from the Department of Human Genetics, University of Pittsburgh Graduate School of Public Health, Pittsburgh, USA. For his PhD, He worked in the fields of DNA repair and telomere biology, both of which have profound implications in aging and cancer. He then pursued postdoctoral training in genomics at the Department of Developmental Biology, University of Pittsburgh, USA. In 2015 he joined Memorial Sloan Kettering Cancer Center (MSKCC), New York, USA, as a research fellow, where he designed and successfully developed a liquid biopsy test for early detection and treatment monitoring of a subset of head and neck cancers associated with Human Papillomavirus infections.
Present work area and expertise
Dr. Damerla’s research interests broadly include cancer genomics and functional characterization of mutations in genes involved in the etiology of genetic disease. He is also interested in studying how disturbances in DNA repair pathways effect tumorigenesis along with developing novel genomics technologies for early detection and treatment monitoring of cancers by liquid biopsy.
He has initiated a biobank with the support of a seed grant from MAHE for storage of tissue, blood and nucleic acid samples from patients with cancer and other genetic disease. Dr. Damerla currently works on the following 3 research projects in collaboration mainly with the Manipal Comprehensive Cancer Care Center and multiple departments from Kasturba Medical College Manipal, Manipal College of Pharmaceutical Sciences and Manipal College of Dental Sciences.
1. Cervical Cancer Genomics: Droplet digital PCR assay to monitor HPV circulating tumor DNA as a biomarker for disease monitoring in patients with cervical cancer. Whole genome sequencing of cervical cancer tumors to identify novel mutations and genetic signatures relevant to the Indian population.
2. Head and Neck Cancer Genomics: prevalence of HPV associated head and neck cancers and the role of HPV in modulating different tumorigenesis pathways in head and neck cancer.
3. Developing a single assay based on long range sequencing for detection of both point mutations and structural variations in a panel of genes frequently mutated in Hereditary Breast and Ovarian Cancers.

Dr. Pavan Agrawal
Assistant Professor,
DBT, India, Ramalingaswami re-entry fellow,
Centre for Molecular Neurosceinces,
Kasturba Medical College,
Manipal Academy of Higher Education,
Manipal, Udupi,
Karnataka, India -576104
Email: pavan.agrawal@manipal.edu
Webpage: https://www.agrawallab.com/
Research Experience
Dr. Pavan Agrawal joined Manipal Academy of Higher Education in October 2019, as an Assistant Professor and Department of Biotechnology– Government of India, Ramalingaswamy re-entry Fellow at the KMC, Manipal’s Centre for Molecular Neurosciences.
Pavan obtained his PhD at the CSIR-Centre for Cellular and Molecular Biology, Hyderabad, India with Prof. L. S. Shashidahara on Drosophila development. He then pursued his Postdoctoral research in Neurobiology at the HHMI, Janelia Research Campus, USA, where he worked with Dr. Ulrike Heberlein and Dr. Loren Looger.
His work identified whole brain as well as cell type specific transcriptional and epigenetic changes in the Drosophila brain due to social isolation stress. Using brainwide RNA-seq screen he identified a neuropeptide Dsk which increases aggressive behavior driven by social isolation. Interestingly, intravenous injection of Dsk’s homologue- CCK, is known to trigger panic attacks in humans and is used for screening anti-anxiety drugs; suggesting evolutionary conservation in mechanisms regulating stress.
His work also led to the development of a method called ‘mini-INTACT’ which enabled epigenetic characterization of rare cell types from the brain. This innovative method reduced the required input tissue material by ~100-fold as compared to earlier methods. Using mini-INTACT he purified dopaminergic neurons and identified shifts in epigenetic and transcriptional landscape mediated by social isolation. Using quantitative behavioral assays, his work identified causal transcriptional mechanisms in dopaminergic neurons that alter sleep behavior driven by social isolation. This work opens the possibility of using Drosophila as a model for cell type specific behavioral epigenetics.
His research program at KMC, Manipal is funded by several competitive research grants including- DBT, India Extramural Research grant; DBT; Ramalingaswami re-entry fellowship, and MAHE intramural seed grant. As Assistant Professor at KMC, Manipal he is advising several PhD candidates, JRFs as well as MBBS students interested in biomedical research.
Present work area and expertise
Dr. Pavan is interested in understanding causal mechanisms by which negative stressors such as social isolation, abused drugs such as nicotine and neurodegenerative disorders such as Parkinson’s disease alter nervous system at the epigenomic and transcriptomic level and alter behavior. His laboratory is engaged in several research projects exploring these themes at several levels including molecules, specific cell types in the brain, individual organism as well as groups of animals.
Given the public health problem posed by stressors such as social isolation and abused drugs it is important to understand molecular mechanisms responsible for these phenomena. To approach this problem his lab is using fruit fly Drosophila melanogaster- a powerful and genetically tractable model organism. Drosophila is a pivotal tool to understand genetic and epigenetic mechanisms involved in various brain disorders.
His work combines use of genomic technologies such as ChIP-seq, RNA-seq, ATAC-seq, biochemistry and quantitative high-throughput behavioral assays including aggression, sleep and courtship; as well as classical Drosophila genetics. He has a keen interested in developing novel molecular tools to investigate nervous system function in cell type specific manner. More details about his work can be found on his webpage: https://www.agrawallab.com/
He is peer reviewer of several international journals including- eLIFE, Genetics, Plos One, G3: Genes, Genomes, Frontiers in Behavioral Neuroscience, Scientific reports and Molecular Neurobiology.
Publications: (*Co/Corresponding author)
1. Kent, C., Agrawal, P*. Regulation of social Stress and neural degeneration by activity-regulated genes and epigenetic mechanisms in dopaminergic neurons. Mol. Neurobiol. 57, 4500–4510 (2020). https://doi.org/10.1007/s12035-020-02037-7
2. Agrawal P*, Kao D, Chung P, Looger LL. The neuropeptide Drosulfakinin regulates social isolation-induced aggression in Drosophila. J Exp Biol. 2020 Jan 29;223:jeb207407. https://doi.org/10.1242/jeb.207407
3. Agrawal, P.*, Chung, P., Heberlein, U. Kent, C. Enabling cell-type-specific behavioral epigenetics in Drosophila: a modified high-yield INTACT method reveals the impact of social environment on the epigenetic landscape in dopaminergic neurons. BMC Biol 17, 30 (2019). https://doi.org/10.1186/s12915-019-0646-4
Collaborations
Dr. Pavan is a firm believer in open and collaborative science and has several active collaborations both nationally and internationally. Some of his national collaborations include researchers from NCCS, Pune; NCBS, Bangalore; M. S. University Baroda; IISER, Pune; IISER, Kolkata etc. His international collaborations include HHMI, Janelia Research Campus, USA; University of California San Diego, USA; Northwestern University, Chicago, USA; York University, Canada etc.
Pavan also enjoys teaching genetics and neuroscience as guest lecturer across several institutions nationwide and is enthusiastically popularizing use of small model organism like fruit fly in medical research. He has a keen interest in interdisciplinary work and collaborates within KMC, MAHE with clinicians as well as bioengineers at Manipal Institute of Technology, MAHE. He is actively engaged in several institutional committees, including a core member of KMC PhD cell, KMC Research Seminar series and ‘Grant Clinics’ which helps researchers across KMC, Manipal with their grant writing efforts.
Dr. Piyush Behari Lal, M.Sc, MS-PhD
Assistant Professor
DBT Ramalingaswami Fellow
Department of Microbiology
Kasturba Medical College
Manipal Academy of Higher Education
Manipal, Udupi, Karnataka
India 576104
Email: piyush.lal@manipal.edu

Research Experience:
He has done M. Sc. in Biomedical Sciences from Bundelkhand University, Jhansi, UP; research internship at ACBR, University of Delhi; MS-PhD from University of Texas at Dallas, TX, USA (Dr. Larry Reitzer’s Lab); and postdoc from Great Lakes Bioenergy Research Center, University of Wisconsin – Madison, WI, USA (Dr. Patricia Kiley’s lab).
Dr. Piyush Behari Lal has more than 14 years of research experience in Genetics, Biochemistry, molecular biology, metabolic engineering and synthetic biology in a model bacterium gamma-proteobacterium Escherichia coli and a non-model alpha-protobacteria Zymomonas mobilis ZM4
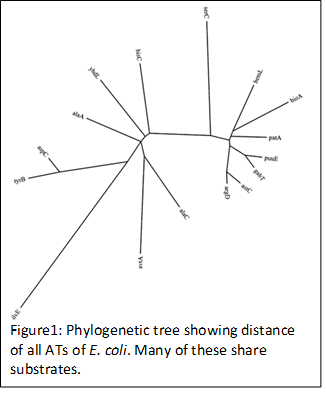
He has two years of research experience in organic synthesis and screening of organic compounds against clinical bacterial isolates at Bundelkhand University and at University of Delhi. In his PhD dissertation, he studied aminotransferase (ATs) redundancy in E. coli (Figure 1) and regulation of Alanine synthesis (Dr. Larry Reitzer’s Lab). Redundancy and promiscuity have always been acknowledged, but the extent of these properties have not been systematically studied. Dr. Piyush identified the ATs of arginine (3 ATs) and lysine (4 ATs) synthesis in E. coli and showed how general such redundancy was. His work revealed that certain ATs are extraordinarily redundant and promiscuous and also that these characteristics are conserved traits in evolution, possibly to protect vital cellular functions during stress.

His postdoctoral research work (Dr. Patricia Kiley’s Lab) involved development of genetic tools for Z. mobilis, improvement of gene annotation and development of Z. mobilis strains for robust biosynthesis of biofuels and industrially relevant byproducts (Figure 2). Z. mobilis. The ethanologenic bacterium Z. mobilis is considered a promising candidate for microbial conversion of plant biomass (lignocellulosic biomass/hydrolysate) into biofuel and other valuable products. Like other non-model bacteria, Z. mobilis has several limitations such as lack of facile genetic tools, genetically tractable strains, poor genome annotation etc. To make strain optimization feasible and efficient, he developed genetic tools for Z. mobilis, identified and removed RM systems of Z. mobilis to improve conjugation efficiency of foreign genes. He also performed research for improvement of genome annotation using Tn-Seq approach, which is a high throughput sequencing-aided transposon mutagenesis approach that can analyze phenotype of several thousand mutant phenotype in one set of experiment. This work was a part of a collaborative project at the Great Lakes Bioenergy Research Center (GLBRC), which is one of the five Bioenergy Research Centers of the Department of Energy.
Present work area and expertise:
Dr. Piyush’s current work involves two major projects: Understanding of virulence factors of Melioidosis causing bacteria Burkholderia pseudomallei (Medical Biotechnology) and developing a better biofuel-producing strain of Zymomonas mobilis (Synthetic Biology).
Melioidosis (Medical Biotechnology): Melioidosis, which is caused by B. pseudomallei, is endemic to south Asia and north Australia and is considered an emerging infectious disease in many tropical developing countries. B. pseudomallei is a highly pathogenic bacterium. It is classified as a category B biological agent by the Centers for Disease Control and Prevention and is considered a global threat because it has a mortality rate of up to 50% even with antibiotic treatment.
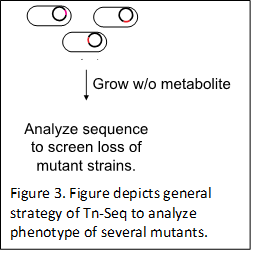
Many virulence genes have been identified in different populations of B. pseudomallei strains isolated from different clinical or environmental isolates around the world. However, no studies have explored the molecular mechanism of pathophysiology of B. pseudomallei strains isolated from Indian patients, which are considerably different from the strains isolated from other parts of the world. This work uses comprehensive approach to understand metabolic capabilities and to improve functional annotation of genome. A high throughput Tn-Seq approach (Figure 3) will be used in this study to analyze phenotype and fitness of several thousand mutant strains (Tn-library). As the data will measured by averaging across multiple independent transposon insertions in each gene, the results will be very robust in systematic analysis of the role of individual genes for pathogenesis and overcoming antibiotic resistance.
Biofuels (Synthetic Biology): Elucidating strategies of resource allocation (Figure 4) and metabolism is crucial for engineering strains with industrial relevance. Dr. Piyush’s lab focuses on metabolic pathways in a natural ethanologen Zymomonas mobilis ZM4. Z. mobilis is an alpha-proteobacteria with several attractive physiological features to become a platform strain for producing valuable products from compounds present in plant biomass. Plant biomass extracts (hydrolysates) contain several carbohydrates and several amino acids, which are not metabolized by Z. mobilis. Dr. Piyush’s lab focuses on equipping this bacterium with novel genes and pathways to channel carbon present in agriculture waste into specially biofuels or industrially relevant byproducts.
Collaborations:
Indian collaborators:
· Dr. Chiranjay Mukhopadhyay,Director, Manipal Institute of Virology, and Professor at Department of Microbiology, KMC, Manipal, Karnataka
· Dr. Somasish Ghosh Dastidar, Assistant Professor and Ramalingaswami Fellow, Center for Molecular and Neuroscience, KMC, Manipal, Karnataka
· Dr. Rama Damerla, Assistant Professor and Ramalingaswami Fellow, KMC, Manipal, Karnataka
· Dr. Ranita Ghosh Dastidar, Assistant Professor, Department of Biochemistry, KMC, Manipal, Karnataka
· Dr. Vandana KE, Professor and Head, Department of Microbiology, KMC, Manipal, Karnataka
· Dr. Dr. Divendu Bhushan, Associate Professor, AIIMS Patna, Bihar
· Dr. Prathyusa K., Associate Professor, AIIMS Patna, Bihar
Foreign collaborators:
· Dr. Larry Reitzer, Professor, University of Texas at Dallas, Richardson, TX, USA.
· Dr. Patricia Kiley, Professor and Chair, Department of Biomolecular Chemistry, University of Wisconsin – Madison, Madison, WI, USA.
· Dr. Tim Donohue, Professor and Director, GLBRC, University of Wisconsin – Madison, Madison, WI, USA.
· Dr. Robert Landick, Professor and Science Director, GLBRC, University of Wisconsin – Madison, Madison, WI, USA.
· Dr. Aseem Ansari, Professor and Chair, St. Jude Children’s Research Hospital, Memphis, USA.

Somasish Ghosh Dastidar, M.Sc., M.S., Ph.D.
Assistant Professor
DBT Ramalingaswami Fellow
Center for Molecular Neurosciences
2nd Floor, Physiology Block
Kasturba Medical College, Manipal
Manipal Academy of Higher Education (MAHE), Manipal
Madhav Nagar; Dist-Udupi. Karnataka, India; Pin :576104
Office :(+91) 08202923614
Cell : (+91) 8088872591
Email: somasish.gd@manipal.edu
Website: https://www.dastidarlab.org/
Research Experience
Dr. Dastidar was born in Eastern India. He did his Bachelors in Microbiology from the University of Pune, and Masters in Biotechnology from Visva Bharati University, India. He did a M.S. in Cell and Molecular biology through his integrated MS-PhD program at the University of Texas at Dallas. Dr. Dastidar did his PhD in the lab of Prof. Santosh D’Mello, where he investigated the role of transcription factors, in the modulation of neuronal survival/apoptosis and the underlying mechanism(s) behind these processes. During his PhD training Somasish gained expertise in neurodegeneration, genetics, cell and molecular biology, biochemistry, and advanced imaging.
After his PhD, Dastidar joined the laboratory of Prof. Albert La Spada who has developed an extensive research program in neurodegeneration and neurotherapeutics, Dr. La Spada is credited for the discovery of trinucleotide repeats in the etiology of Spino Bulbar Muscular Atropy, the first in the series of many repeat nucleotide disorders.
During his postdoctoral training Dastidar built on his foundation, to study miRNA expression & regulation patterns, generation/characterization of sophisticated transgenic or knock in mice models, mice behavioural and histopathological analysis, stereotactic surgery in mice, lentiviral technology, stem cell modelling, CRISPR/Cas9 based genome editing and a host of cell biological processes such as autophagy, stress granule biology, unfolded protein response, proteasomal machinery, nucleolar stress critical for cell homeostasis He uses animal cell lines, primary neuronal cells, primary glial cells, induced Pluripotent Stem cells, mice and rats as model systems to test his scientific hypothesis.
Somasish has published in Cell Metabolism, Acta Neuropathologica, Nature Neuroscience, Journal of Neuroscience, Journal of Biological Chemistry, Journal of Clinical Investigation, eLife, Cell reports, GeroScience etc. Somasish has mentored over 30 undergraduate and graduate students during his PhD and postdoctoral training. Dastidar enjoys mentoring budding scientists and is very passionate about it. Currently he is mentoring PhD students, Masters Student and MBBS student. The Dastidar Lab is thankful to DBT, ICMR and MAHE for their generous funding.
Present work area and expertise
The Dastidar lab focuses on solving puzzles involving molecular mechanisms behind the etiology of age-related disorders (such as Parkinson’s & ALS) with a commitment to identify therapeutic targets. The labs in collaboration with the Dr. Chiranjay Mukhopadhyay, is interested to determine the pathogenesis behind Melioidosis which is caused by Burkholderia pseudomallei. The lab utilizes novel molecular and cell biology tools and technique, biochemistry, in vitro and in vivo model systems, mice genetics, regenerative biology, microbiology, NGS and CRISPR/Cas9 based functional genomics screening approaches.
We are currently focusing on the following projects:
Role of Reduced protein translation in neurodegenerative conditions:
As reduced protein synthesis may decrease stress on cellular protein quality control systems in the cell, the overall goal of this project is to examine the role of reduced protein translation in neuroprotection, by testing if increased expression of 4EBP1 can ameliorate toxicity in a series of in vitro and in vivo models of PD.
The outcome from this project will not only confirm the importance of reduced protein translation in neuroprotection, but, if successful, will also seek to establish the mechanistic pathways underlying neuroprotection promoted by reduced protein translation. This work will thus have therapeutic implications for developing treatments for debilitating neurodegenerative disorders (such as AD and ALS) and possibly other mitochondrial diseases.
To identify activators and repressors of mammalian eukaryotic Initiation Factor 4 Binding Proteins (4EBPs) as potential therapeutic targets.
The overarching goal of this project is the characterization of the eIF4E-BPs with a focus on identifying their upstream activators or repressors which may represent as therapeutic targets for treatment in cancer of the CNS and neuroinflammatory disorders. This work will thus have therapeutic implications for developing treatments for PD and possibly other pathologically similar neuroinflammatory disorders.
Identify genetic targets or mini chaperones to alleviate the molecular pathophysiology of Amyotrophic Lateral Sclerosis.
Small molecules, peptides, molecular chaperones and antisense oligos are currently being pursued worldwide as therapeutic avenues against proteinopathies. We are studying if mini-chaperones can be protective against proteinopathies and ALS. Furthermore, we are also studying the molecular mechanisms behind the etiology of ALS4 caused due to mutations in the Senataxin gene.
Deciphering the underlying mechanisms and basis of pathophysiology of Neuromelioidosis, a rare but fatal emerging tropical disease in India.
Melioidosis is a life-threatening neglected tropical disease caused by soil saprophyte Burkholderia pseudomallei. India with the highest burden have an expected mortality rate of over 30,000 per year. Curated data (Kasturba Hospital) from Melioidosis cases with confirmed CNS involvement show the incidence of Neuromelioidosis to be comparatively higher in the coastal region of Karnataka, with meningitis as the prominent presentation. Neuromelioidosis has a high mortality rate. Critical gap in understanding the host-pathogen interaction, bacteria entry across the blood-brain-barrier, evasion of host immune response, virulence factors, and genetic heterogeneity of Burkholderia requires immediate attention. Comorbidities such as long-standing uncontrolled diabetes mellitus and chronic kidney disease aggravates Neuromelioidosis. No diagnostic kit or vaccine is available commercially. High prevalence of diabetes in the Indian population and B. pseudomallei being a Tier one select agent with intrinsic multidrug resistance are significant concerns of national relevance.
We collaborate with national and international scientists to understand the molecular mechanisms behind this disease using cutting edge technology involving microbiology, biochemistry, genetics, molecular and cell biology, animal models, iPSCs, CRISPR/Cas9 based functional genomics screening approaches and “omics” based approaches.
Collaborations:
Dr. Dastidar have collaborators in University of California Irvine, Boise State University, University of Missourie-School of Medicine & University of Washington, Seattle (USA); Deakin University (Australia); Pontifícia Universidade Católica do Paraná (Brazil); Kasturba Medical College, Manipal College of Health Professions, Manipal Institute of regenerative Medicine, Manipal School of Life Sciences (MAHE, India); National Institute of Biomedical Genomics; Indian Association for the Cultivation of Science; National Institute of Technology Karnataka, Surathkal; Indian Institute of Technology- Mandi; Visva Bharati University and University of Calcutta, (India).
